Deep learning for prediction of mechanism in acute ischemic stroke using brain diffusion magnetic resonance image
Article information
Abstract
Background
Acute ischemic stroke is a disease with multiple etiologies. Therefore, identifying the mechanism of acute ischemic stroke is fundamental to its treatment and secondary prevention. The Trial of Org 10172 in Acute Stroke Treatment classification is currently the most widely used system, but it often has a limitations of classifying unknown causes and inadequate inter-rater reliability. Therefore, we attempted to develop a three-dimensional (3D)-convolutional neural network (CNN)-based algorithm for stroke lesion segmentation and subtype classification using only the diffusion and apparent diffusion coefficient information of patients with acute ischemic stroke.
Methods
This study included 2,251 patients with acute ischemic stroke who visited our hospital between February 2013 and July 2019.
Results
The segmentation model for lesion segmentation in the training set achieved a Dice score of 0.843±0.009. The subtype classification model achieved an average accuracy of 81.9%, with accuracies of 81.6% for large artery atherosclerosis, 86.8% for cardioembolism, 72.9% for small vessel occlusion, and 86.3% for control.
Conclusion
We developed a model to predict the mechanism of cerebral infarction using diffusion magnetic resonance imaging, which has great potential for identifying diffusion lesion segmentation and stroke subtype classification. As deep learning systems are gradually developing, they are becoming useful in clinical practice and applications.
INTRODUCTION
Acute ischemic stroke has various causes based on the causative mechanism, consisting of large artery atherosclerosis (LAA), cardioembolism (CE), small vessel disease, stroke of other determined etiologies, or stroke of undetermined etiology. Classification of acute ischemic stroke based on the cause is important for treatment and secondary prevention. The most widely used classification system is the Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification [1,2]. However, this classification method shows moderate inter-rater reliability in classifying acute ischemic stroke and has a limitation of frequently classifying strokes as having an undetermined etiology [3]. To overcome this limitation, efforts are underway to develop a computerized algorithm for acute stroke diagnosis; however, these have not shown sufficient results [4,5].
Various diagnostic methods such as brain imaging and heart tests are required to determine the causative mechanisms of acute ischemic stroke. Early diagnosis of the stroke subtype using this classification system can positively affect treatment, prognosis, and secondary prevention [1]. Diffusion-weighted magnetic resonance imaging (MRI) is widely used to diagnose acute stroke. It has superior performance in detecting hyperacute lesions and very small ischemic lesions and in distinguishing chronic and acute lesions [6] compared to brain computed tomography (CT) and conventional MRI. Furthermore, simultaneous use of the apparent diffusion coefficient (ADC) map and diffusion-weighted imaging (DWI) allow for more accurate distinction of the lesion of acute ischemic stroke, providing important information about the time window of the lesion [7]. The diffusion imaging lesion pattern, which provides useful information for the early diagnosis of acute ischemic stroke, has been reported to be closely related to the stroke subtype [8,9].
Various deep learning algorithms based on convolutional neural networks (CNNs) have been proposed for diagnosing acute ischemic stroke in brain MRI images [10-21]. These studies have shown that deep learning can detect stroke lesions more accurately than traditional machine-learning techniques and can extract meaningful features for severity evaluation or prognosis prediction. Researchers have proposed lesion segmentation techniques for patients with acute ischemic stroke based on the U-Net architecture [16,17]. To efficiently exploit the contextual information of volumetric MRI data, Zhang et al. [18] proposed a stroke lesion segmentation technique using a three-dimensional (3D) fully connected-DenseNet. Although the aforementioned studies demonstrated that deep learning can classify patients with acute ischemic stroke via lesion segmentation, a classification technique for predicting the treatment mechanism of acute ischemic stroke has yet to be reported. In this study, we presented a 3D CNN-based model for stroke lesion segmentation and subtype classification using only DWI and ADC images from patients with acute ischemic stroke.
METHODS
Study population
The participants were 2,251 patients with acute ischemic stroke who visited our hospital between February 2013 and July 2019. Information on acute ischemic stroke was compiled from a registry. All patients with acute ischemic stroke were reviewed by at least three stroke specialists and classified according to the TOAST classification. There were 1,789 patients with LAA, CE, and small vessel occlusion (SVO), excluding stroke of other determined etiologies or stroke of undetermined etiology. Among them, 1,396 patients underwent DWI and ADC (Fig. 1). There were 608 patients with LAA, 441 with CE, and 359 with SVO. Among the healthy patients who visited the hospital during the same period, 400 who showed normal MRI findings at our clinic were included as controls. The patients’ sex, age, National Institutes of Health Stroke Scale score, and medical history, including stroke, hypertension, diabetes mellitus, and atrial fibrillation, are listed in Table 1. In the control group, brain images without clinical information were used. Baseline characteristics were presented as frequencies (percentages). Continuous variables with normal distributions are presented as means±standard deviation, whereas variables with non-normal distributions are presented as medians (interquartile ranges).
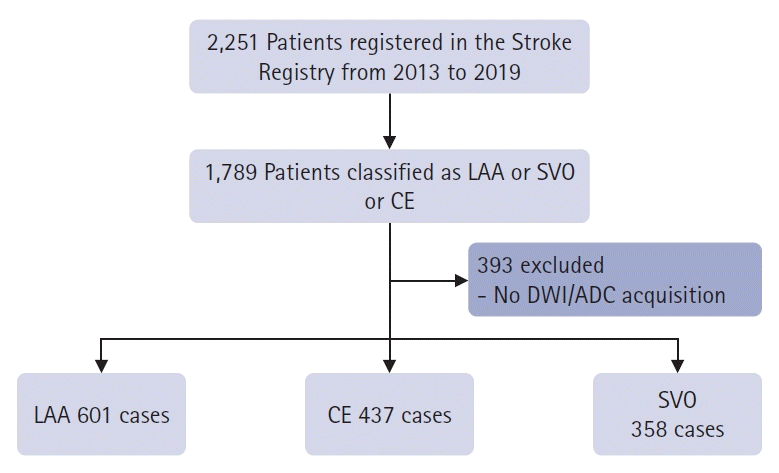
Study profile. LAA, large artery atherosclerosis; SVO, small vessel occlusion; CE, cardioembolism; DWI, diffusion-weighted imaging; ADC, apparent diffusion coefficient; AI, artificial intelligence.
Imaging acquisition
MRI was performed using various machines including 1.5 T (Achieva, Philips Healthcare) and 3.0 T (Ingenia CX, Philips Healthcare; Achieva, Philips Healthcare) scanners. The parameters of the DWI sequence were as follows: repetition time, 2,500–3,000 ms; echo time, 80 ms; slice thickness, 3–5 mm; intersection gap, 1 mm; field of view, 220×220 mm; matrix size, 256×256 (approximately 2×2 mm in-plane resolution); and b values, 0 and 1,000 sec/mm2. Each apparent ADC map was generated automatically using the manufacturer’s software.
Data preparation
To produce a “ground truth” reference standard for training and evaluating the subtype classification model, each patient was classified into four classes (LAA, SVO, CE, and Control) according to the TOAST classification system. Lesion areas in each DWI slide were manually annotated by two experienced neurologists using in-house annotation software. Finally, each lesion was cross-validated and labelled, with a final decision agreed upon by both raters. To address the data distribution and validation methodology in our study, we adopted a 5-fold Stratified Cross-Validation approach. This ensures the proportionality of the class distribution across each fold, which is crucial for maintaining the integrity of the validation process given the imbalanced nature of our dataset. The preprocessing pipeline was meticulously designed to normalize and standardize the MRI images obtained from various vendors with different acquisition parameters. Each patient's MRI data comprising varying numbers of slices were resampled to a uniform 3D voxel size of 256 (H) × 256 (W) × 128 (D). This resampling is pivotal for aligning the spatial dimensions across all the datasets. To address the intensity variations due to different magnetic resonance (MR) parameters and scanner calibrations, we performed intensity normalization using the window center and width values provided in the digital Imaging and communication in medicine (DICOM) file metadata. This step adjusts the pixel intensity values to a standard scale, thereby enhancing the image comparability. Additionally, DWI, which inherently have varying numbers of slices owing to different scanning protocols, were standardized by selecting a fixed number of slices that best represented the essential features required for accurate segmentation. This uniform pre-processing approach ensures that subsequent segmentation algorithms operate on data that reflect consistent anatomical structures and tissue characteristics, thereby enabling a more reliable and valid comparative analysis across all images. Practically, for each of the five folds, we allocated 60% of the data for training purposes, 20% for validation, and 20% for testing. This division was performed independently within each fold to confirm the generalizability and reliability of the model's performance.
Lesion segmentation mode
Our segmentation model, based on a 3D CNN called V-Net [22] is illustrated in Fig. 2. The model consists of an encoder that extracts feature maps from local 3D volumes and a decoder that predicts stroke lesions using feature maps. Because our model has a very deep architecture, we employed a residual block to alleviate the gradient vanishing and exploding problems. The residual block contains (1) a 3D convolutional layer with kernel size 3×3×3 and (2) a residual skip connection and two 3D convolutional layers with kernel size 3×3×3, each following batch normalization and a rectified linear unit, respectively. In the network encoder, residual blocks were utilized for feature extraction and max-pooling layers, with a stride of two to reduce spectral dimensionality. In contrast, the decoder consists of up-convolutional layers with strides of two, followed by residual blocks after feature-map concatenation. Skip connections from the layers of equal resolution in the encoder provide high-resolution features to the decoder. A sigmoid activation layer was connected to the last layer of the decoder to calculate a probability map of the stroke lesions.
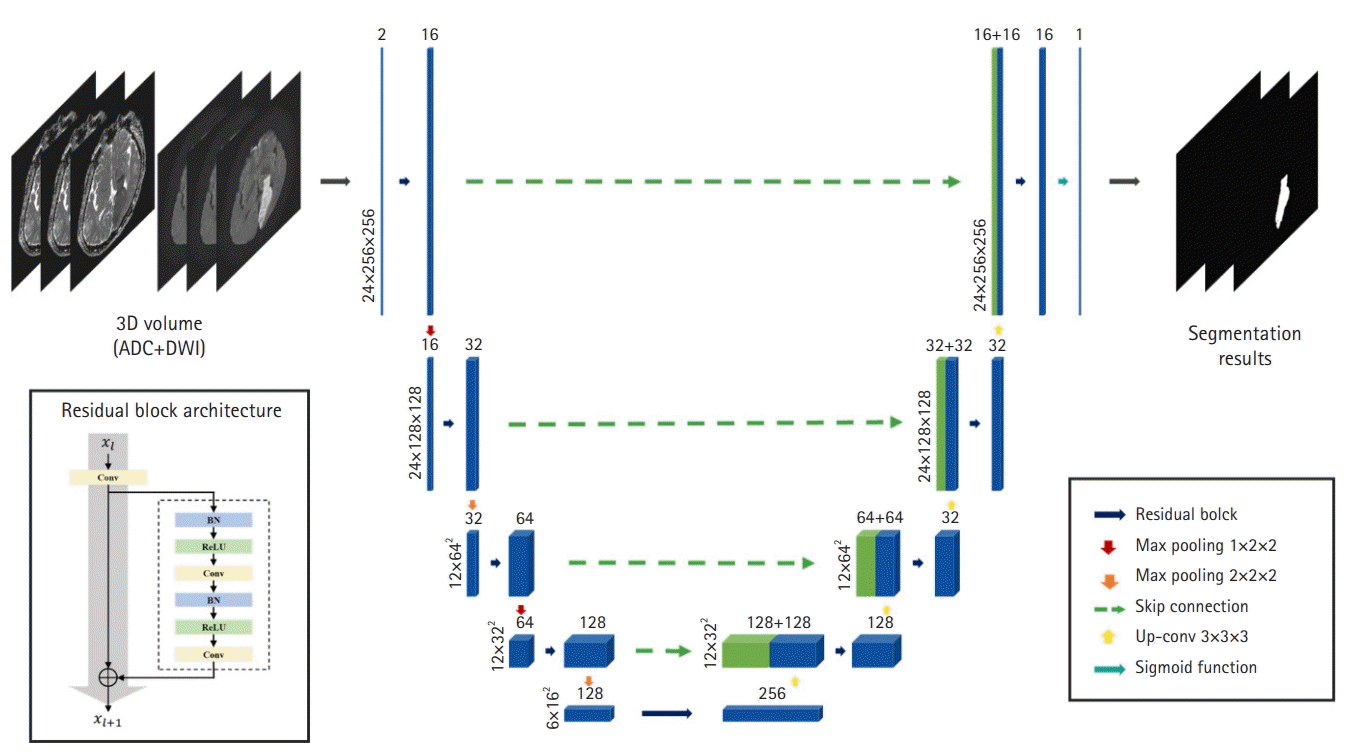
Our network architecture for stroke lesion segmentation. Based on three-dimensional (3D) U-Net, the network learns the features based on a hierarchy framework starting from simple features such as edges and shapes to high-level features in the deeper levels. ADC, apparent diffusion coefficient; DWI, diffusion-weighted imaging; Conv, convolution; BN, batch normalization; ReLU, rectified linear unit; Up-conv, up-convolutional.
The model was trained over 200 epochs with an Adam optimizer, an initial learning rate of 1e-5, and a batch size of 8. The model was trained from scratch without using pretrained weights. We tested various loss functions such as weighted cross-entropy loss, L1 loss, and Dice loss; Dice loss achieved the best performance. Furthermore, to address the data scarcity problem, data augmentation techniques such as rigid transformation, horizontal/vertical flip, Gaussian noise, and gamma correction were randomly triggered in each training session.
Subtype classification model
As illustrated in Fig. 3, our classification model predicted the probabilities of the four classes: LAA, SVO, CE, and Control. For feature extraction, we adopted a residual block in the lesion-segmentation model. In addition, to guide the network to focus on the lesions, feature maps were enhanced using the lesion prediction results provided by the segmentation model. Specifically, the enhanced feature map Fenh is obtained by
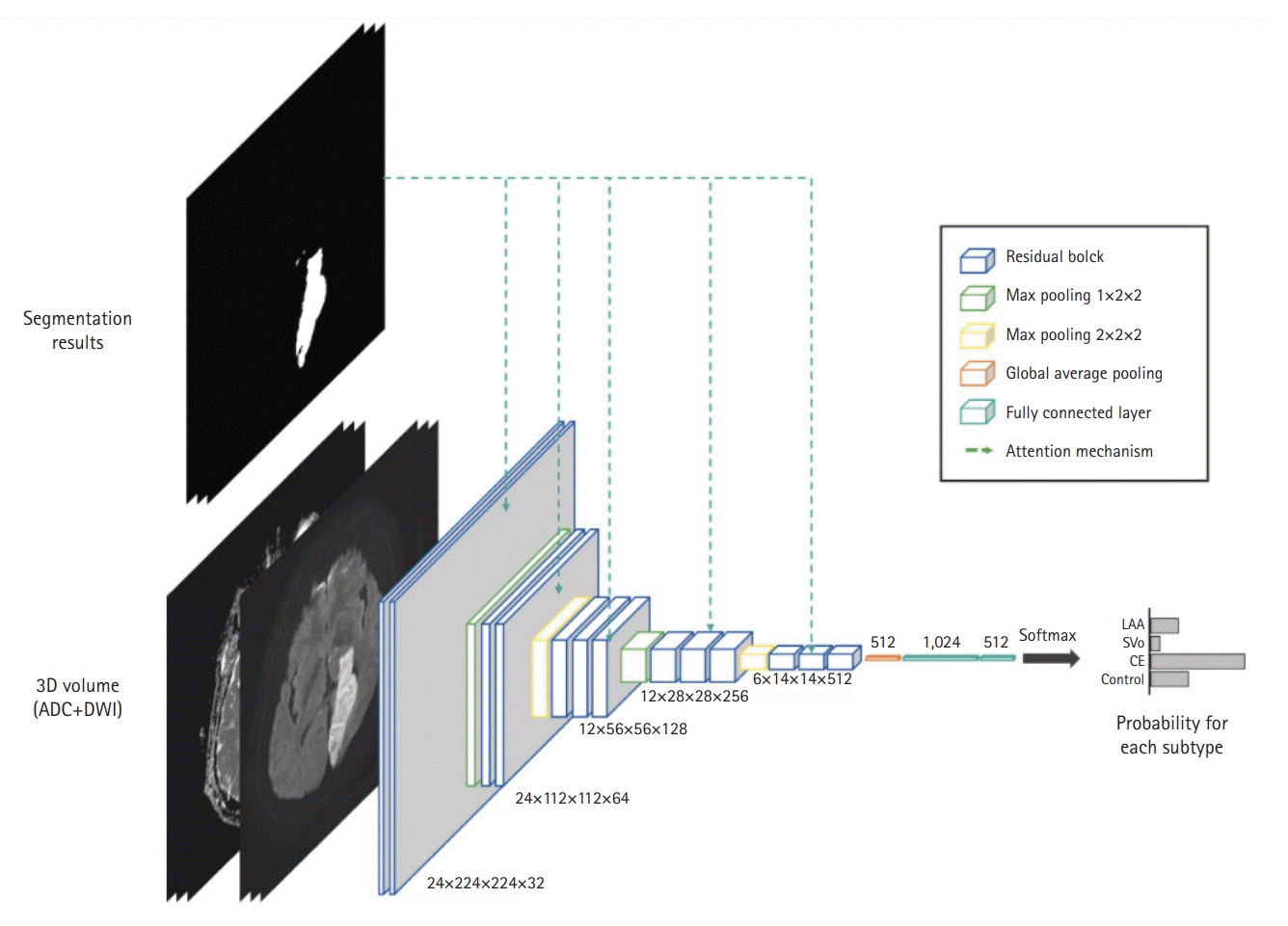
Our network architecture for stroke subtype classification. To guide the network towards the lesion areas, we adopted the attention mechanism using the lesion segmentation result. ADC, apparent diffusion coefficient; DWI, diffusion-weighted imaging; LAA, large artery atherosclerosis; SVO, small vessel occlusion; CE, cardioembolism.
Fenh = F × (1 + h(A)), (1)
where F and A are the feature maps extracted by each residual block and lesion segmentation result, respectively. h(·) is a bilinear interpolation to match the spatial resolutions between F and A. This attention mechanism significantly improves the classification performance of the model by guiding the network to focus on lesion areas to predict stroke subtypes. The model was trained over 400 epochs with an Adam optimizer, an initial learning rate of 1e-5, and a batch size of 4. Categorical cross-entropy loss was utilized, and the model was trained from scratch. We used a data-augmentation technique to train the classification model.
RESULTS
Lesion segmentation model
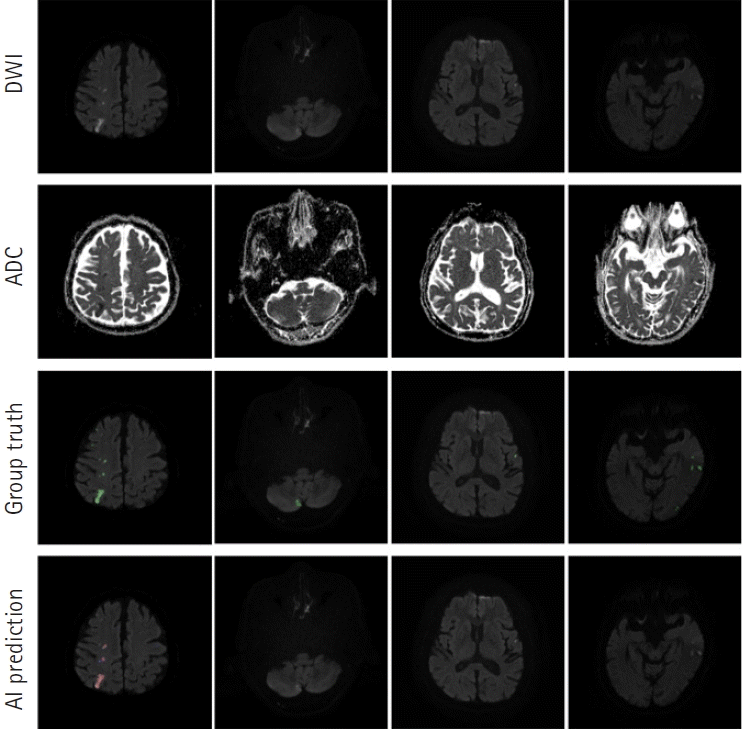
Our segmentation model for lesion segmentation in the training set achieved a Dice score of 0.891± 0.034. For the test set, our model resulted in a Dice score, precision, and recall of 0.843±0.009, 0.842±0.012, and 0.844±0.017, respectively. Fig. 4 shows some examples of lesion prediction results compared with assessments by neurologists. Our segmentation model accurately predicted extremely small lesions. Most cases of failure occurred when the lesions had very poor contrast in the diffusion images, as shown in Fig. 5.
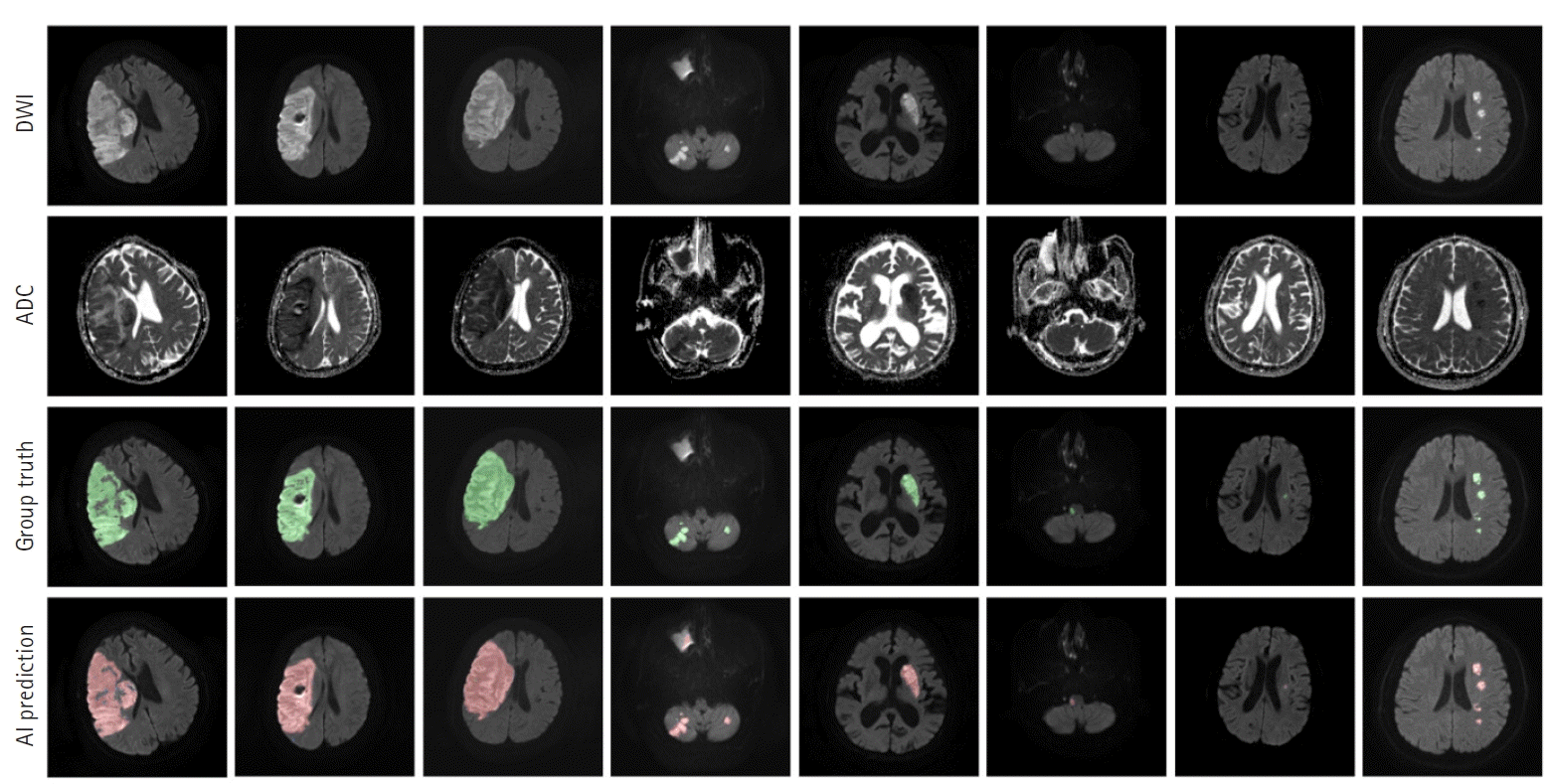
Prediction outcomes using our lesion segmentation model. In each panel, the images in the first and second rows are diffusion-weighted imaging (DWI) and apparent diffusion coefficient (ADC) slices, respectively. The third-row images are the “ground truth” labels annotated by two neurologists, while the fourth-row images show lesion areas predicted by our model. AI, artificial intelligence.
Stroke subtype classification model
To underscore the benefits of leveraging segmentation data, we integrated an enhanced feature map into our stroke-subtype classification model. By applying an enhanced feature map informed by the segmentation results, the model obtains the spatial context that the raw images lack. This context allows the model to “see” beyond mere pixel intensity, recognizing patterns and structures pertinent to stroke subtype.
This strategic modification led to a notable improvement in the performance metrics. Before integrating the segmentation information, the average classification accuracy of the model is 71.1%. However, with the incorporation of an enhanced feature map, the accuracy significantly increased to 81.9%. This effect was evident across all subtypes, with accuracies of 81.6% for LAA, 86.8% for CE, 72.9% for SVO, and 86.3% for control. The enhanced feature map sharpens the model's ability to concentrate on lesion-specific areas, thereby refining the differentiation process between various stroke subtypes.
Fig. 6 shows the confusion matrix obtained using the subtype classification model. Our model showed lower accuracy for SVO than for other subtypes, indicating that the model confused SVO with control cases owing to its poor analysis performance for small lesions.
DISCUSSION
This is the first study to perform subtype classification of stroke mechanisms by analyzing the patterns of acute ischemic stroke lesions through deep learning based on a 3D-CNN using DWI and ADC in patients with acute ischemic stroke. The main findings of this study are as follows. First, the 3D-CNN-based segmentation accuracy for acute ischemic stroke lesions was 0.843 based on the Dice score. Second, in terms of subtyping to classify the cause of acute ischemic stroke, the predicted degree of cause classification according to the TOAST classification, which is the "ground truth," was confirmed to be 81.3% for LAA, 84.6% for SVO, and 73.0% for CE.
With technological advances, brain imaging plays a crucial role in diagnosing and identifying mechanisms underlying the development of acute ischemic stroke according to technological advances [23]. Among the various MR sequences, DWI and ADC maps are useful tools for the early detection of acute ischemic lesions and for differentiating between stroke mimics and acute ischemic stroke [24]. Several previous studies have attempted to segment the infarction volume in acute ischemic stroke using artificial intelligence (AI). Various imaging patterns of acute ischemic stroke in DWI lesions correlate with pathogenic mechanisms. In the case of cardiac embolic stroke, acute stroke lesions on DWI often show single cortical/subcortical lesions or occur multiple times in various vascular branches. Multiple unilateral lesions in the anterior circulation are characteristic findings of arteriogenic embolism. Meanwhile, small infarction (2–20 mm in diameter) lesions observed in the deep acute ischemic white matter, basal ganglia, thalamus, and pons were highly associated with SVO [8,25]. We attempted to apply a doctor’s diagnostic process to determine the cause of acute ischemic stroke based on the characteristic findings of brain MRI using AI.
However, there are many limitations in predicting the pathogenesis of acute ischemic stroke using only DWI/ADC maps. The TOAST classification system is the most widely used system for classifying acute ischemic stroke based on its pathogenesis. Clinical findings and the results of ancillary diagnostic studies, including brain imaging and cardiac evaluation, were used to classify patients’ acute ischemic stroke mechanism [1]. Although widely use and popular, its overall inter-rater agreement is moderate. Its reliability is notably lower for small-vessel occlusion and strokes of undetermined causes, especially when compared to LAA and CE [3]. To overcome this, an improved classification method that applies a new diagnostic technique was used; nevertheless, it still has limitations [4,5,26,27]. In particular, acute ischemic stroke with unknown mechanisms, such as LAA, CE, SVO, and stroke of other determined etiologies, is known as cryptogenic stroke. It is observed in approximately all the patients with acute ischemic stroke [28]. These cryptogenic strokes are often observed as embolic strokes, and are called embolic strokes of undetermined source (ESUS) [29]. There is a need to determine the mechanism of ESUS and provide proper treatment; however, a definitive method for achieving this objective remains elusive [30]. We conducted this study to diagnose acute ischemic stroke using a deep learning algorithm. To the best of our knowledge, this model is the first algorithm for identifying the mechanism of cerebral infarction in patients with ESUS. In the future, it will be necessary to create a multimodal algorithm that includes cerebrovascular imaging, laboratory data, and cardiac tests, such as transthoracic echocardiography, transesophageal echocardiography, and electrocardiography. We expect to improve the model in this study.
Our study introduces a novel approach to lesion segmentation and stroke subtype classification that significantly advances technology beyond previous methodologies [10-21]. Unlike traditional techniques [16,17] which process individual slides and therefore cannot utilize contextual information from adjacent slides, often resulting in diminished accuracy, our technique leverages a 3D CNN with a deep residual network architecture. This allowed for stable learning and improved recognition of complex patterns across multiple slides, culminating in a high Dice score of 0.845 in the test set.
Moreover, our subtype classification model exhibited an average accuracy of 81.9%, which is a notable improvement over the existing models. This is achieved through an attention mechanism that utilizes the lesion information predicted by the segmentation model, focusing on key areas for accurate prediction. This not only identified the stroke subtype but also highlighted the specific regions the model analyzed to arrive at its conclusion.
The integration of these advanced segmentation and classification models is expected to have a substantial impact on medical AI applications that rely on 3D volumetric data, such as CT and MRI scans. Our approach sets a new precedent for accuracy and reliability in medical diagnostics, offering a comprehensive solution that outperforms previous single-slice-based techniques.
This study has several limitations. First, the labeling of subtypes in the stroke prediction is unclear. Despite our meticulous process of employing the TOAST classification system and expert annotations by two experienced neurologists, the subjective nature of clinical diagnoses presents the potential for inconsistency. The difficulty in standardizing labels across different raters and cases is an inherent limitation not only in our study, but also in the broader context of machine learning applications in stroke subtype classification. This could result in variability, affecting the reliability of our model. Recognizing this limitation, we emphasize the need for continuous improvement in annotation methodologies and exploration of more objective measures in future studies to minimize such discrepancies. Second, this study lacks external validation. Therefore, there may be a bias in this model, and it is necessary to improve it by performing external validation in future studies. Third, this algorithm does not include images of cerebral infarction caused by causes other than LAA, CE, or SVO. To predict and diagnose these mechanisms, additional clinical data, such as cerebrovascular imaging, laboratory studies, and cardiological evaluation, are required in addition to DWI/ADC. In this study, the algorithm was only trained on three mechanisms that are known to be diagnosable or predictable by DWI patterns. Therefore, it is limited in classifying cerebral infarction caused by other mechanisms. In our follow-up study, we plan to improve the algorithm by including various types of clinical data. Fourth, this study is a case-control study. The study was based on a stroke database from a single center; hence, there is a possibility of selection bias in the selection of subjects. Therefore, it is necessary to overcome this limitation using multicenter data in subsequent studies. Finally, this study was conducted using data collected from a single cohort, which limited our consideration of variations in the MR parameters. Therefore, the generalizability of our findings to datasets with different MR parameters may be limited. Specifically, our normalization approach based on Window Center and WindowWidth may not be applicable to other datasets with varying imaging protocols. This limitation highlights the need for further research using diverse MRI datasets to validate and refine our methodology. Future studies should aim to incorporate data from multiple sources with varying MR parameters to ensure broader applicability and robustness of the findings.
In summary, this study aimed to predict the pathogenesis of cerebral infarction using only brain diffusion MRI and apply it clinically. Using only the initial diffusion MRI information, we present a feasible model that predicts the mechanism of occurrence by applying an algorithm based on a 3D-CNN through deep learning. The diffusion lesion volume measurement and stroke subtype classification using our proposed method showed a strong correlation with those performed by manual segmentation and subtype classification by professional neurologists. This study is significant because it is the first to predict the mechanism of acute ischemic stroke by using diffusion MRI alone. In future studies, it will be necessary to develop a multimodal algorithm that includes not only diffusion MRI, but also other brain imaging modalities and clinical data to predict the exact pathogenesis of cerebral infarction.
Notes
Ethics statement
This single-center, retrospective case-control study was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Institutional Review Board (No. 2023-10-017), which waived the requirement for informed consent.
Conflict of interest
Moon Ku Han and Jeong-Ho Hong are editorial board members of the journal, but they were not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by funding from the Academic Research Program of the Chungbuk National University in 2022.
Author contributions
Conceptualization: all authors. Methodology: BKK, SP, DIL, KSY. Formal analysis: BKK, SP, DIL, KSY. Data curation: BKK, SP, DIL, KY. Visualization: BKK, SP, DIL, KSY. Project administration: BKK, SP, DIL, KSY. Funding acquisition: KSY. Writing–original draft: BKK, SP, DIL, KY. Writing–review & editing: all authors.